
Hi there! I have put together a python package for analyzing single cell
BCR/TCR data from 10x Genomics 5’ solution! It streamlines the
pre-processing, leveraging some tools from immcantation suite, and
integrates with scanpy/anndata for single-cell BCR/TCR analysis. It also
includes a couple of functions for visualization. Try it out on !
Also check out our review at Nature Methods on “Single-cell immune repertoire analysis”:
Irac et al. (2021), Single-cell immune repertoire analysis, Nature Methods.
Overview
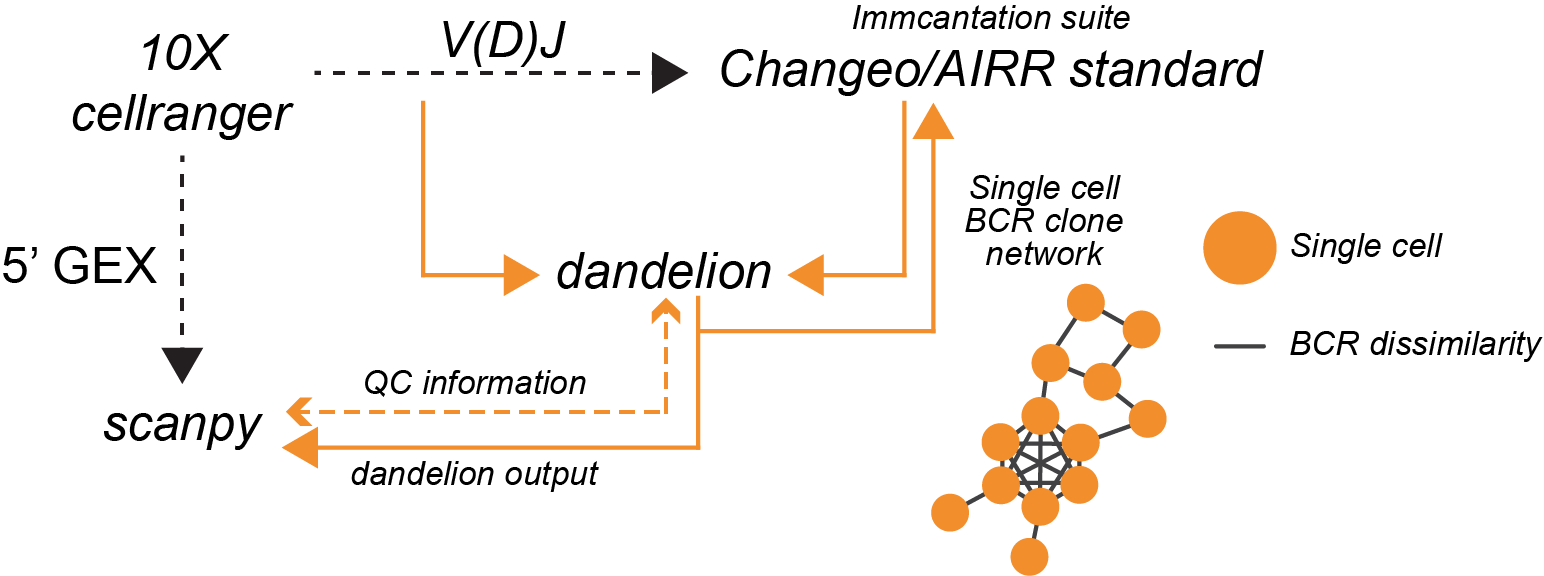
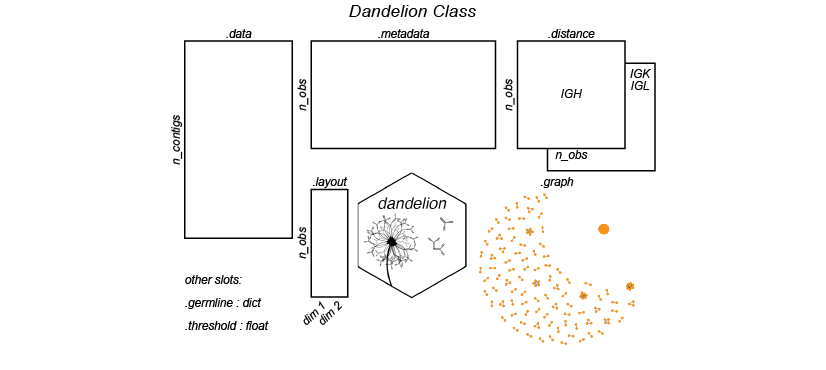
Illustration of the Dandelion class slots
Please refer to the documentation or the notebooks here:
The raw files for the examples can be downloaded from 10X’s Single Cell Immune Profiling datasets website.
Installation
Singularity container
dandelion now comes ready in the form of a singularity container
which has all the required dependencies installed:
singularity pull library://kt16/default/sc-dandelion:latest
singularity shell --writable-tmpfs -B $PWD sc-dandelion_latest.sif
This will load up a conda-environment that has all the required dependencies installed.
This can be used for the preprocessing steps by navigating to the data folder and use:
singularity run -B $PWD sc-dandelion_latest.sif dandelion-preprocess
Python package
Start off by creating a conda environment containing scanpy, following official scanpy instructions. Once done, run the following:
conda install -c conda-forge graph-tool
pip install sc-dandelion
Between this and the pipelines within the singularity container, you should be covered for most of your needs.
Basic requirements
Python packages
# conda
python>=3.7 (conda-forge)
numpy>=1.18.4 (conda-forge)
pandas>=1.0.3 (conda-forge)
distance>=0.1.3 (conda-forge)
jupyter (conda-forge) # if running via a notebook
scikit-learn>=0.23.0 (conda-forge)
numba>=0.48.0 (conda-forge)
pytables>=3.6.1 (conda-forge)
seaborn>=0.10.1 (conda-forge)
leidenalg>=0.8.0 (conda-forge)
plotnine>=0.6.0 (conda-forge)
graph-tool>=2.3.5 (conda-forge) # optional
# Other executables (through conda)
blast>=2.10.1 (bioconda)
igblast>=1.15.0 (bioconda)
# pip
anndata>=0.7.1
scanpy>=1.4.6
scrublet>=0.2.1
changeo>=1.0.0
presto>=0.6.0
polyleven>=0.5
networkx>=2.4
rpy2>=3.4.2
Acknowledgements
I would like to acknowledge the contributions from Dr. Chenqu Suo, Dr. Krysztof Polanksi, Dr. Sarah Teichmann and Prof. Menna Clatworthy, who helped with the initial conception of the project and for all discussions.
I would also like to acknowledge Dr. Ondrej Suschanek, Dr. Benjamin Stewart, Dr. Rachel Bashford-Rogers, Dr. Jongeun Park, Dr. Cecilia-Dominguez Conde, Dr. Kirsten Stewart, Dr. Hamish King and Dr. Peng He with whom I have had very useful discussions. I would also like to thank my wife who helped name the package, because she thought the plots looked like a dandelion =D.
Support
Support is provided on a voluntary basis, as time permits.
If there are any ideas, comments, suggestions, thing you would like to know more etc., please feel free to email me at z.tuong@uq.edu.au or post in the issue tracker and I will get back to you.
Citation
Please also cite the following paper if you use version 0.3.0 onwards:
Suo et al. (2023), Dandelion uses the single-cell adaptive immune receptor repertoire to explore lymphocyte developmental origins, Nature Biotechnology.
Chenqu Suo, Krzysztof Polanski, Emma Dann, Rik GH Lindeboom, Roser Vilarrasa-Blasi, Roser Vento-Tormo, Muzlifah Haniffa, Kerstin B Meyer, Lisa M Dratva, Zewen Kelvin Tuong, Menna R Clatworthy, Sarah A Teichmann. Dandelion uses single cell adaptive immune receptor repertoire to explore lymphocyte developmental origins. Nature Biotechnology 2023.04.13; doi: https://doi.org/10.1038/s41587-023-01734-7*
dandelion was originally published in:
Stephenson et al. (2021), Single-cell multi-omics analysis of the immune response in COVID-19, Nature Medicine.
Emily Stephenson, Gary Reynolds, Rachel A Botting, Fernando J Calero-Nieto, Michael Morgan, Zewen Kelvin Tuong, Karsten Bach, Waradon Sungnak, Kaylee B Worlock, Masahiro Yoshida, Natsuhiko Kumasaka, Katarzyna Kania, Justin Engelbert, Bayanne Olabi, Jarmila Stremenova Spegarova, Nicola K Wilson, Nicole Mende, Laura Jardine, Louis CS Gardner, Issac Goh, Dave Horsfall, Jim McGrath, Simone Webb, Michael W Mather, Rik GH Lindeboom, Emma Dann, Ni Huang, Krzysztof Polanski, Elena Prigmore, Florian Gothe, Jonathan Scott, Rebecca P Payne, Kenneth F Baker, Aidan T Hanrath, Ina CD Schim van der Loeff, Andrew S Barr, Amada Sanchez-Gonzalez, Laura Bergamaschi, Federica Mescia, Josephine L Barnes, Eliz Kilich, Angus de Wilton, Anita Saigal, Aarash Saleh, Sam M Janes, Claire M Smith, Nusayhah Gopee, Caroline Wilson, Paul Coupland, Jonathan M Coxhead, Vladimir Y Kiselev, Stijn van Dongen, Jaume Bacardit, Hamish W King, Anthony J Rostron, A John Simpson, Sophie Hambleton, Elisa Laurenti, Paul A Lyons, Kerstin B Meyer, Marko Z Nikolic, Christopher JA Duncan, Ken Smith, Sarah A Teichmann, Menna R Clatworthy, John C Marioni, Berthold Gottgens, Muzlifah Haniffa. Single-cell multi-omics analysis of the immune response in COVID-19. Nature Medicine 2021.04.20; doi: https://dx.doi.org/10.1038/s41591-021-01329-2
If you use the pre-processing tools/functions, please cite the relevant manuscripts from the immcantation suite, including:
Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356-8 (2015). doi: https://doi.org/10.1093/bioinformatics/btv359
Gadala-Maria D, Yaari G, Uduman M, Kleinstein SH. Automated analysis of high-throughput B cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proceedings of the National Academy of Sciency of the United States of America, E862-70.
References
Bashford-Rogers et al. (2013), Network properties derived from deep sequencing of human B-cell receptor repertoires delineate B-cell populations, Genome Research.
Bashford-Rogers et al. (2019), Analysis of the B cell receptor repertoire in six immune-mediated diseases, Nature.
Dann et al. (2022), Differential abundance testing on single-cell data using k-nearest neighbor graphs, Nature Biotechnology. GitHub.
Gadala-Maria et al. (2015), Automated analysis of high-throughput B cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles, Proceedings of the National Academy of Sciency of the United States of America.
Gupta et al. (2015), Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data, Bioinformatics.
Irac et al. (2024), Single-cell immune repertoire analysis, Nature Methods.
Setty et al. (2019) Characterization of cell fate probabilities in single-cell data with Palantir, Nature Biotechnology. GitHub.
Sleckman et al. (1998) Assembly of productive T cell receptor delta variable region genes exhibits allelic inclusion, Journal of Experimental Medicine.
Stephenson et al. (2021), Single-cell multi-omics analysis of the immune response in COVID-19, Nature Medicine.
Sturm et al. (2020), Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data, Bioinformatics. GitHub.
Suo et al. (2022), Single cell antigen receptor analysis reveals lymphocyte developmental origins, bioRxiv.
Suo et al. (2023), Dandelion uses the single-cell adaptive immune receptor repertoire to explore lymphocyte developmental origins, Nature Biotechnology.
Wolf et al. (2018), Scanpy: large-scale single-cell gene expression data analysis, Genome Biology. GitHub.
